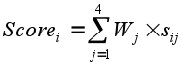José Luis Oliveira
Universidade de Aveiro, DETI / IEETA
3810-193 Aveiro, Portugal
jlo@ua.pt
(+351) 234 370 500
What is QuExT?
QuExT (Query Expansion Tool) is a document indexing and retrieval application that obtains, from the MEDLINE database, a ranked list of publications that are most significant to a particular set of genes. Document retrieval and ranking are based on a concept-based methodology that broadens the resulting set of documents to include documents focusing on these gene-related concepts. Each gene in the input list is expanded to its various synonyms and to a network of biologically associated terms. Currently, the expansion is based on proteins, metabolic pathways and diseases (this last one only when the selected organism is Homo sapiens). The retrieved documents are ranked according to user-definable weights for each of these concept classes. By simply changing these weights, users can alter the order of the documents, allowing them to obtain for example, documents that are more focused on the metabolic pathways in which the initial genes are involved, rather than on the genes themselves.
How does it work?
QuExT receives as input a list of genes and a corresponding organism. The gene list can be typed into the input box or uploaded in a text file. Genes can be separated by commas or spaces. The organism to consider is selected from the drop-box menu. Figure 1 shows the query expansion procedure.
When the user submits the form, gene names or identifiers in the input are checked against a database and mapped to an internal identifier corresponding to the selected organism. Genes which are not found in the database are rejected from further analysis.
QuExT then creates an expanded query and searches a local index of the PubMed database for documents matching this query.
Query expansion is performed as follows: for each gene in the query, the algorithm obtains, from a term expansion table corresponding to the selected organism, all the alternative gene, protein, pathway and disease names corresponding to that gene’s internal ID. The full list of terms from all input genes is then accumulated in four separate query strings (one for each concept type). Each term obtained from expanding all genes is used to search the index.
QuExT runs four index searches using the four query strings obtained in the query expansion stage (one for each concept type). For each search, the documents that match the query and the corresponding scores are obtained. Resulting documents and corresponding scores are kept on separate lists, one for each concept class.
Notice that while the term expansion takes into account the selected organism, to avoid going from a gene in one organism to a related term in a different organism, this is not true for document retrieval. Since the indexing does not distinguish between different species referred in the articles, a search for a gene name in H. sapiens may return results referring to the same gene but in mice, for example.
Finally, the results from the document retrieval stage are assembled and documents are re-ranked in terms of the defined weights for each concept. The final score for document i is obtained as a weighted sum of the four concept-based scores:
where Wj is the weight attributed to the concept type j and sij represents the score for document i in terms of the jth concept type.
References
S. Matos, J. P. Arrais, J. Maia-Rodrigues, J. L. Oliveira, “Concept-based query expansion for retrieving gene related publications from MEDLINE”, BMC Bioinformatics, Apr 28; 11:212, 2010.