José Luis Oliveira
Universidade de Aveiro, DETI / IEETA
3810-193 Aveiro, Portugal
jlo@ua.pt
(+351) 234 370 500
Genomic Name Server
The integration of heterogeneous data sources has been a fundamental problem in database research over the last two decades. The goal is to achieve better methods to combine data residing at different sources, under different schemas and with different formats in order to provide the user with a unified view of the data. Although simple in principle, due to several constrains, this is a very challenging task where both the academic and the commercial communities have been working and proposing several solutions that span a wide range of fields. However, the limitations found on most solutions reflect the difficulty to obtain a simple but comprehensive schema able to accommodate the heterogeneity of the biological domain while maintaining an acceptable level of performance: GeNS is our proposal towards solving this issue.
Installing and using GeNS
The Genomic Name Server can be either downloaded and installed on a local computer or accessed by Web Services. Please keep in mind that GeNS currently requires over 10 GB of disk space and this figure is likely to increase in the near future. Therefore, if disk space is a serious restriction you should consider using the available Web Services. We are currently using
Microsoft SQL Server 2008 but GeNS can be set up in any other DBMS.
a) Setting up a local instance of GeNS
- Download either the full backup of the database (here) or a dump of all the tables (available here): Last update: 24/11/09
- Once inside your DMBS, simply restore the full backup of the database (this is for MS SQL Server 2008 only; a step-by-step walkthrough can be found here) or import the data from the tables to the database.
- Congratulations! GeNS is now ready to be used.
b) Using the Web Services
The Web Services are now available here. Furthermore, a detailed description is also available here (Updated March 24).The Web Services API is in an early stage of development and, as such, users should bear in mind that certains problems may arise during it’s usage.
Advantages
- Easy to understand and use
- Flexible and scalable
- Efficient
- Accessible by several methods
- Improves the cross-database low identifier coverage issue
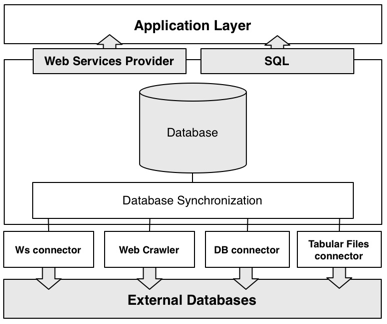
Architecture
GeNS uses four distinct methods for gathering data from external databases: by Web Services, web crawlers, database connectors and finally by tabular files connectors. All of the recovered data is subsquently processed and synchronized to our database. Finally, the data can be accessed via Web Services or by downloading, installing and querying the data with SQL.
Currently, GeNS is importing data from four major databases: UniProt (SwissProt and TrEMBL), KEGG, EMBL – EBI and Entrez. Since these databases already incorporate data from third-party databases, we have over 460.000 unique genes, more than 100.000 biological relations and a hundred and forty distinct datatypes.
Database
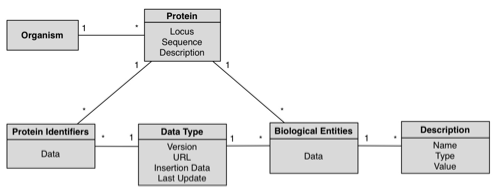
GeNS database was designed with simplicity and extensibility in mind; the following schema is a complete representation of the database.
Concepts:
- Organism: An individual form of life capable of growing, metabolizing nutrients, and usually reproducing. Organisms can be unicellular or multicellular. The Organism table stores taxonomic information; each entry corresponds to an organism with any given number of associated proteins. This table is the root of the hierarchical model. For each organism, we store its scientific and short names.
- Protein: Any of a group of complex organic macromolecules that contain carbon, hydrogen, oxygen, nitrogen, and usually sulfur and are composed of one or more chains of amino acids. The Protein table is where the proteins’ internal identifiers and gene locus are stored; each entry in this table has a referring organism (in which this protein is found) and may have any number of associated biological entities and/or equivalent external databases’ protein identifiers in the ProteinIdentifier and BioEntity tables.
- ProteinIdentifier: The table in which the mapping between the external databases’ protein identifier and BioPortal’s
internal identifier is made. - BioEntity: A table that stores all the biological entities associated with a given protein; this includes,
among other things, pathways and gene ontologies. - DataType: A table listing all the possible external databases from which the biological data may come from; each entry in the ProteinIdentifier and BioEntity tables references this
table, so that we may easily determine the nature (and source) of the
data.
Reproducing the results
The following files allow anyone to reproduce the obtained results regarding the cross-database low identifier coverage issue and the
performance testing queries. You will need a working copy of GeNS in order to use these scripts.