José Luis Oliveira
Universidade de Aveiro, DETI / IEETA
3810-193 Aveiro, Portugal
jlo@ua.pt
(+351) 234 370 500
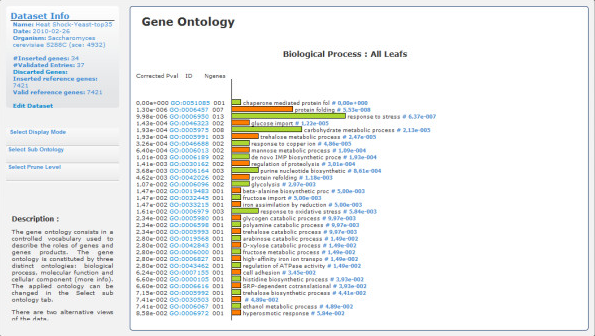
GeneBrowser is a web-based tool that, for a given list of genes, combines data from several public databases with visualisation and analysis methods to help identify the most relevant and common biological characteristics. The functionalities provided include the following: a central point with the most relevant biological information for each inserted gene; a list of the most related papers in PubMed and gene expression studies in ArrayExpress; and an extended approach to functional analysis applied to Gene Ontology, homologies, gene chromosomal localisation and pathways.
Although GeneBrowser can be used to answer many different biological questions, a particular question set was used to tune its development:
- What public databases provide relevant information about my dataset and how can I navigate through them?
- What biological processes are enriched with respect to my input list of genes?
- What are the most relevant metabolic pathways that contain my genes?
- What are the genomic regions of these genes?
- Which are the most relevant homologue classes in my list of genes?
- What gene expression experiments have been previously conducted with the same genes?
- What are the most relevant publications associated with my study?
Feedback
We highly appreciate any feedback you can provide regarding GeneBrowser. jpa@ua.pt. Thank you.
Reference
J. Arrais, J. Fernandes, J. Pereira and J. L. Oliveira, Exploring and identifying common biological traits in a set of genes, BMC Bioinformatics, BMC Bioinformatics 2010, 11:212 (link)