José Luis Oliveira
Universidade de Aveiro, DETI / IEETA
3810-193 Aveiro, Portugal
jlo@ua.pt
(+351) 234 370 500

Problem
The recognition of named entities is a crucial initial task of biomedical text mining. A number of NER solutions have been proposed in recent years, taking advantage of different resources and/or techniques. Currently, the best results are achieved by combining the output of different systems. However, little effort has been spent in such harmonisation solutions, being specific to a corpus and/or non-knowledge based.
Features
- Knowledge-based harmonisation
- Correct, remove and create annotations
- Support several biomedical domains and organisms
- On-demand harmonisation
- Support both NER and normalisation systems
- Automated scripts for simple usage
- Java library for advanced users
- Input and Output in IeXML format
Method
Totum is a innovative harmonisation solution based on Conditional Random Fields, which were trained on several manually curated corpora. Thus, we avoid the single corpus dependency, supporting several biomedical domains and organisms. In the end, Totum harmonises gene/protein annotations provided by several heterogeneous NER solutions, following the gold standard requirements.
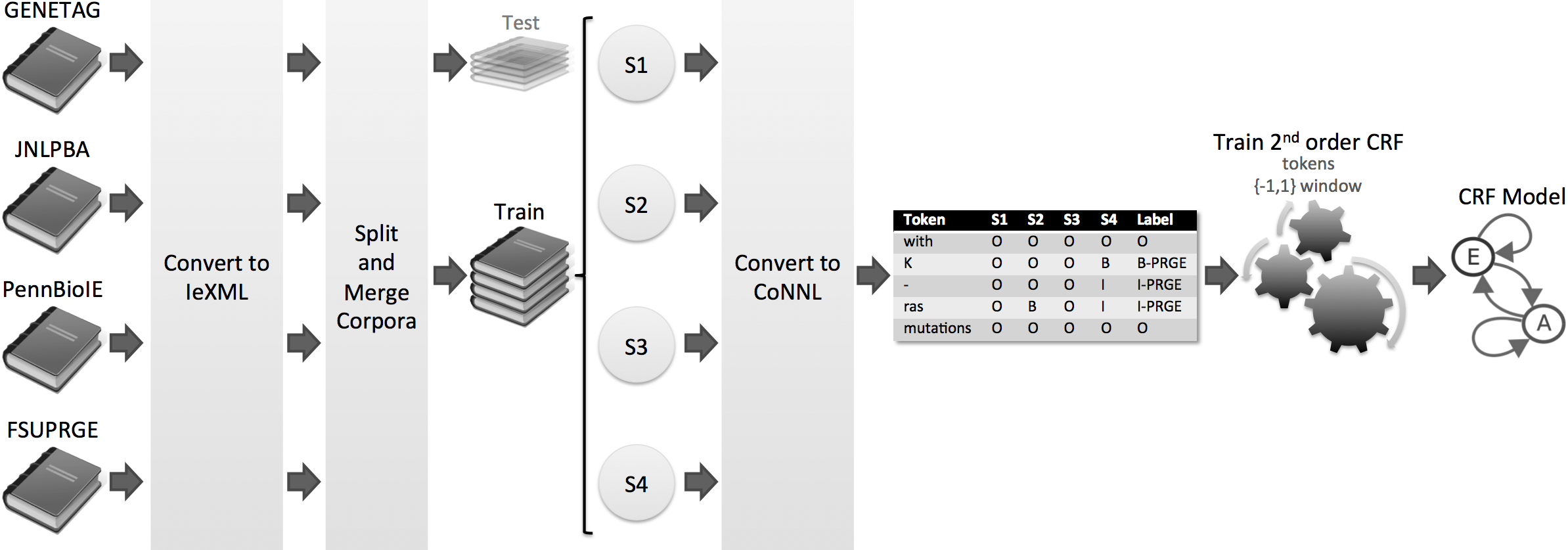
Results
Considering a corpus that contains the test parts of the four corpora, the experiments show that Totum improves the F-measure of state-of-the-art tagging solutions by up to 10% in exact alignment and 22% in nested alignment. Finally, Totum achieves an F-measure of 70% (exact matching) and 82% (nested matching) against the same corpus.
Used tools
- MALLET: framework for statistical natural language processing, providing a Conditional Random Fields implementation;
- Apache OpenNLP: tokenisation and respective model;
- IeXML: annotation guidelines and associated library;
- monq.jfa: fast and flexible text filtering with regular expressions.
Publication(s)
- David Campos, Sérgio Matos, Ian Lewin, José Luís Oliveira, Dietrich Rebholz-Schuhmann. Harmonisation of gene/protein annotations: towards a gold standard MEDLINE. Bioinformatics, vol. 28, no. 9, p. 1253-1261, March 2012. doi:10.1093/bioinformatics/bts125
Team
- David Campos, david.campos(at)ua.pt
- Sérgio Matos, aleixomatos(at)ua.pt
- Ian Lewin, lewin(at)ebi.ac.uk
- José Luís Oliveira, jlo(at)ua.pt
- Dietrich Rebholz-Schuhmann, rebholz(at)ebi.ac.uk
