José Luis Oliveira
Universidade de Aveiro, DETI / IEETA
3810-193 Aveiro, Portugal
jlo@ua.pt
(+351) 234 370 500
Gimli

Annotation of biomedical entity names
the best open-source solution
 Open Source
Open Source- Use, change and distribute
- Social development
 High Performance*
High Performance*- BioCreative: 87,54%
- JNLPBA: 73,05%
 High-end Techniques
High-end Techniques- Linguistic dependency parsing
- Model combination
 Flexible and Scalable
Flexible and Scalable- Extensible architecture
- Fast annotation
 Easy to Use
Easy to Use- Automated scripts
- Java library
* Overall F-measure results achieved using the evaluation methods of the respective challenges.
About
Goal:
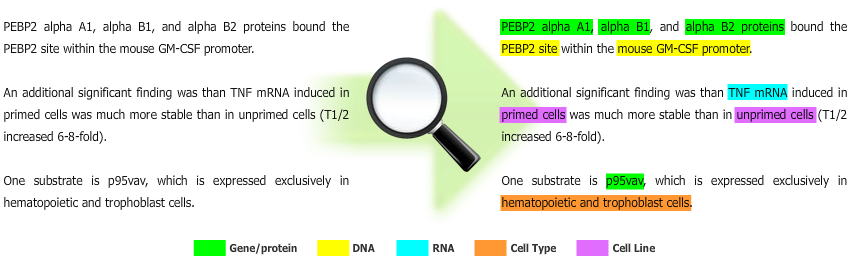
Gimli is a machine learning-based solution for biomedical Named Entity Recognition (NER), which goal is to automatically extract names of biomedical entities from scientific text documents. Currently, Gimli supports the recognition of gene/protein, DNA, RNA, cell line and cell type names.
In summary, Gimli receives raw text as input, and provides text with specific annotations as output.
Methods:
- Machine Learning: Conditional Random Fields (CRFs);
- Features: orthographic, morphological, linguistic parsing and conjunctions;
- Combination: combination of models with different orders and parsing directions;
- Post-processing: parentheses correction and abbreviation resolution.
Publication(s):
- David Campos, Sérgio Matos, José Luís Oliveira. Gimli: open source and high-performance biomedical name recognition. BMC Bioinformatics, vol. 14, no. 1, p. 54, February 2013
Download
Tool:
Get the latest official release of Gimli.
Source code:
Get a copy of the project using the following git command:
Documentation
Full documentation:
Complete information about alternative downloads, installation and usage.
API Javadoc:
Detailed classes, methods and propreties description.
Join us
We have several ideas to make Gimli the most complete and efficient tool for biomedical information extraction. You are welcome to join us and contribute to the development of new and improved features. Please contact us:
 david.campos(a)ua.pt
david.campos(a)ua.ptTeam
- David Campos, david.campos(at)ua.pt
- Sérgio Matos, aleixomatos(at)ua.pt
- José Luís Oliveira, jlo(at)ua.pt


